What is the sequence editor tool in SciNote?
The sequence editor tool in SciNote is an integration of the open-source version of the Open Vector Editor (OVE) developed by Teselagen Biotechnology.
The sequence editor is a molecular biology tool add-on feature integrated into SciNote that allows users to visualize and manipulate genomic files.
You can create a new sequence file or import .dna, .fasta, .gb, and .json files
The tool can be accessed from the Protocol section and the Results section in a Task in SciNote.
To access the tool from the Protocol section:
- Open a Task and navigate to the protocol section.
- Create a new Step if there isn't one already there.
- Click on 'Insert content', select 'File', and then select 'New sequence'.
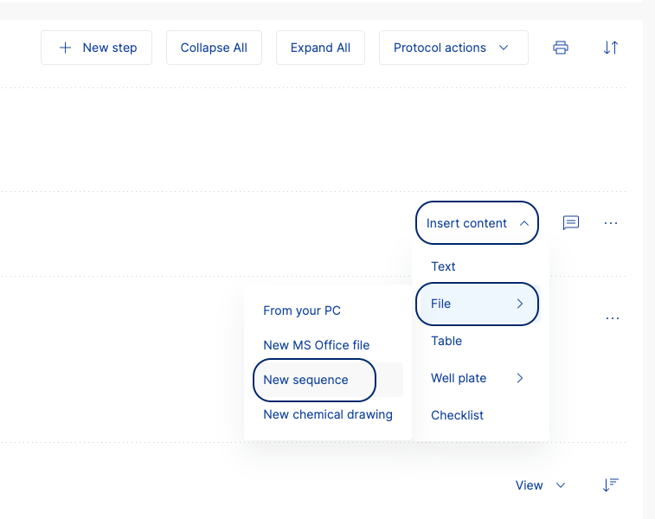
- This will open the sequence editor tool in SciNote.
To access the tool from the Results section:
- Open a Task and navigate to the Results section.
- Add a new Result.
- Click on 'Insert content', select 'File', and then select 'New sequence'.
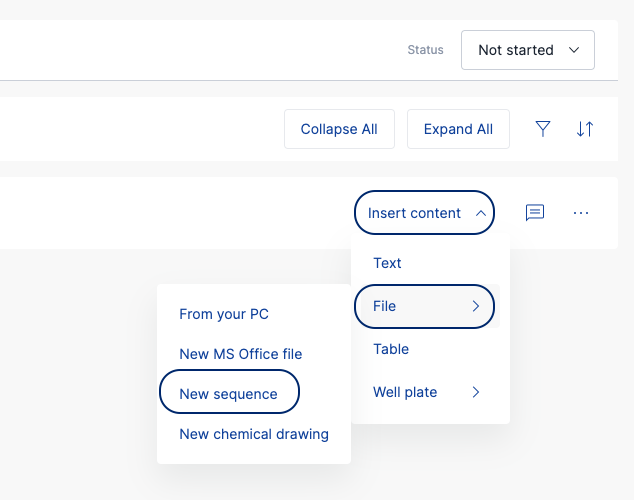
- This will open the sequence editor tool in SciNote.
-
- .json
- .fasta
- .gb
The sequence editor is equipped with various features such as:
- Plasmid visualization
- Primer design
- PCR simulation
- Restriction digestion simulation
...and more!
For more information on this tool, please check out the detailed walkthrough below:
*Reach out to your Customer Success Manager or Account Manager to discuss add-on pricing and enabling the gene sequence editor tool for your SciNote account.
If you have any additional questions, please, do not hesitate to contact us at support@scinote.net. For more information about the Premium plans, please request a quote.